
Microbial Ecology offers articles of original research in full paper and note formats, as well as brief reviews and topical position papers.Ĭoverage includes the ecology of microorganisms in natural and engineered environments genomic and molecular advances in understanding of microbial interactions and phylogeny microbial drivers of biogeochemical processes inter- and intraspecific microbial communication ecological studies of animal, plant and insect microbiology and disease microbial processes and interactions in extreme or unusual environments microbial population and community ecology, and more. Microbial Ecology provides a dedicated international forum for the presentation of high-quality scientific investigations of how microorganisms interact with their biotic and abiotic environments, with each other as well as with their neighbors and hosts, to carry out their diverse functions. Microbial ecology lies at the heart of functioning for almost every ecosystem on the planet, from the deep-sea vents and subsurface systems to human and animal well-being from pristine marine and terrestrial environments to industrial bioreactor functioning. You can use the cat command to quickly print file content to the standard output in the terminal or alternatively concatenate the output. The cat command is a simple and extremely useful command for viewing file contents on Linux.
#LINUX SEQUENCHER FILES HOW TO#
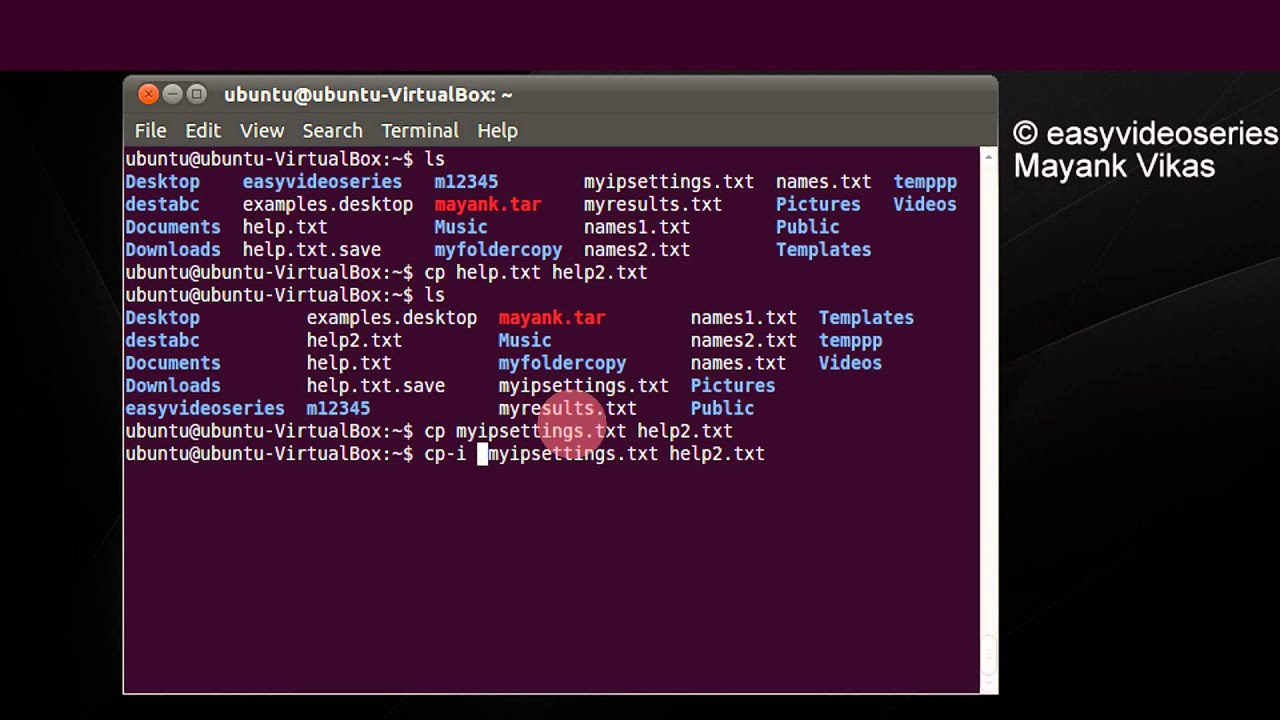
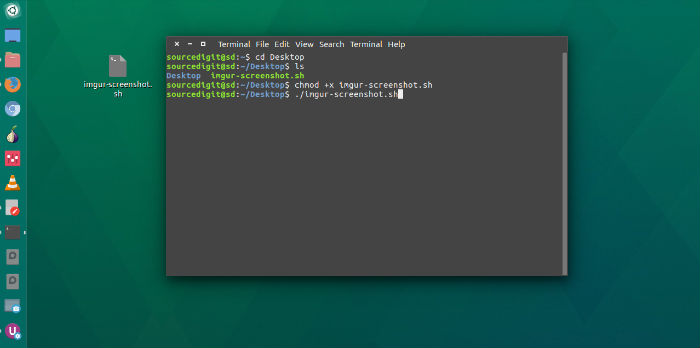
The STITCH software is freely accessible over the Internet at. This tutorial will show you how to view these kinds of files in the Linux terminal. Herein, we describe a software package, search, trim, identify, track, and capture the uniqueness of 16S rRNAs using public and in-house database (STITCH), which offers automated sequence pair splicing and genetic identification, thus simplifying the computationally intensive analysis of large sequencing libraries. In particular, the approach presents two computational challenges: (1) the assembly of a composite sequence from the two partial reads and (2) the subsequent appropriate identification of the organism represented by the newly sequenced clones. The sequencing process typically utilizes a forward and reverse primer pair to produce two partial reads (~700 to 800 base pairs each) that overlap and in total cover a large region of the full 16S rRNA sequence (~1.5 generates very large numbers of 16S rRNA datasets that can overwhelm manual processing efforts leading to both delays and errors. In environmental studies, polymerase chain reaction amplification of 16S rRNA followed by cloning and sequencing of numerous individual clones is an extensively used molecular method for elucidating microbial diversity. A comparison of variable regions within the 16S rRNA gene is widely used to characterize relationships between bacteria and to identify phylogenetic affiliation of unknown bacteria.


 0 kommentar(er)
0 kommentar(er)
